Read and consensus accuracy from Guppy v2.2.3 for a variety of genomes... | Download Scientific Diagram

Consensus decoding improves sequencing accuracy. Reads were run through... | Download Scientific Diagram
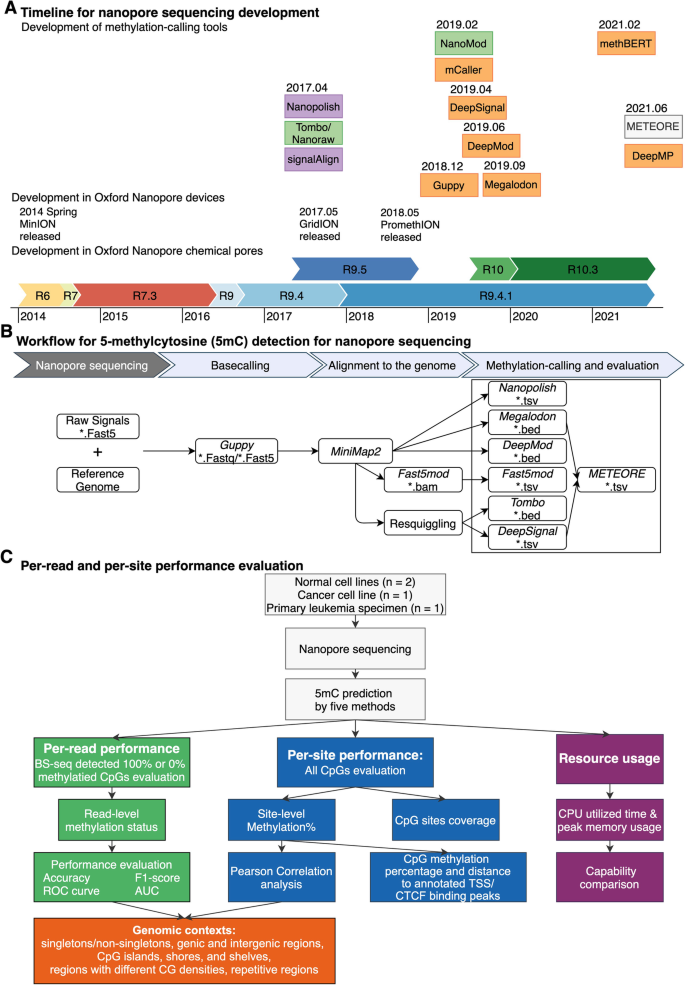
DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation | Genome Biology | Full Text

Frontiers | MasterOfPores: A Workflow for the Analysis of Oxford Nanopore Direct RNA Sequencing Datasets

Recent advances in the detection of base modifications using the Nanopore sequencer | Journal of Human Genetics

Beyond sequencing: machine learning algorithms extract biology hidden in Nanopore signal data: Trends in Genetics
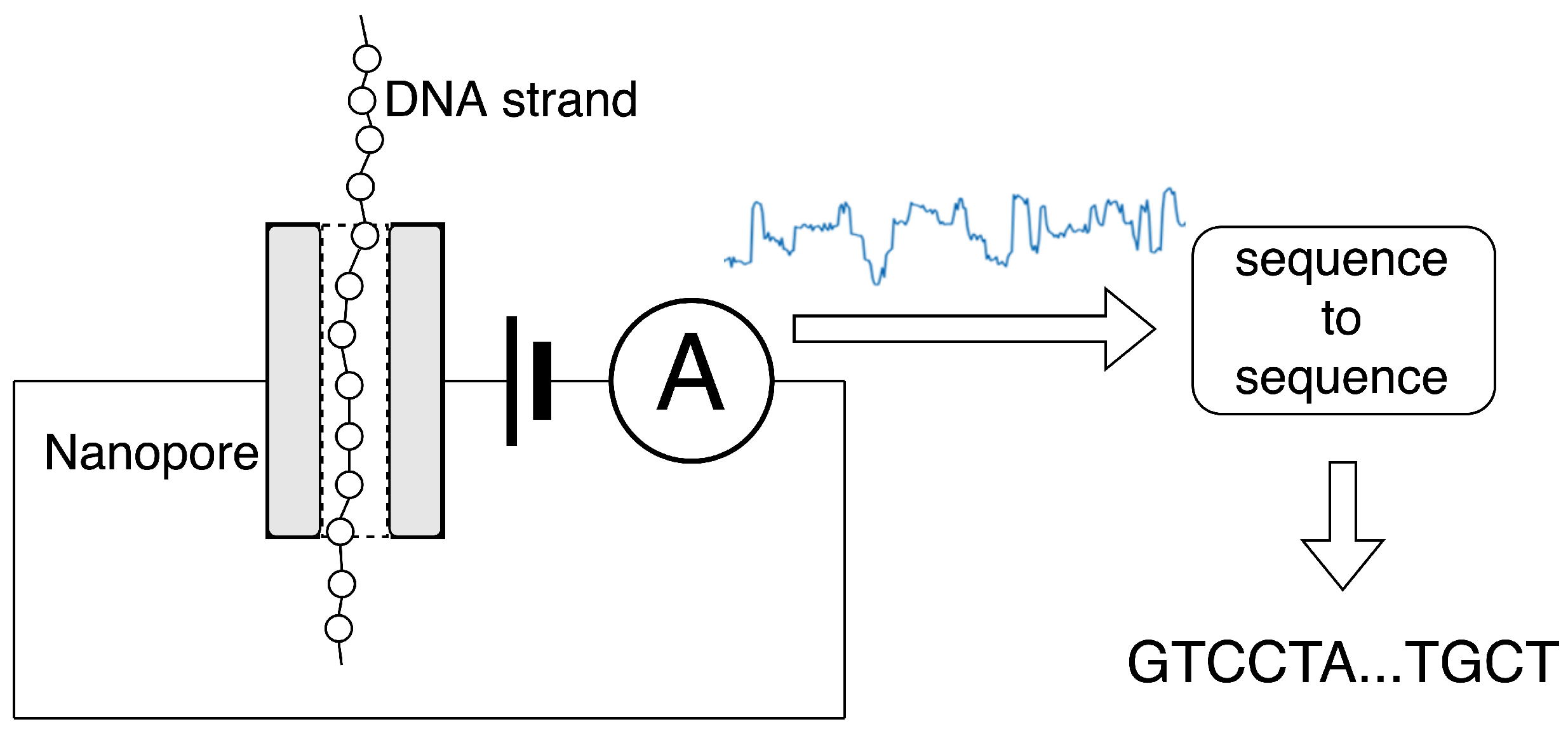
Sensors | Free Full-Text | Basecalling Using Joint Raw and Event Nanopore Data Sequence-to-Sequence Processing
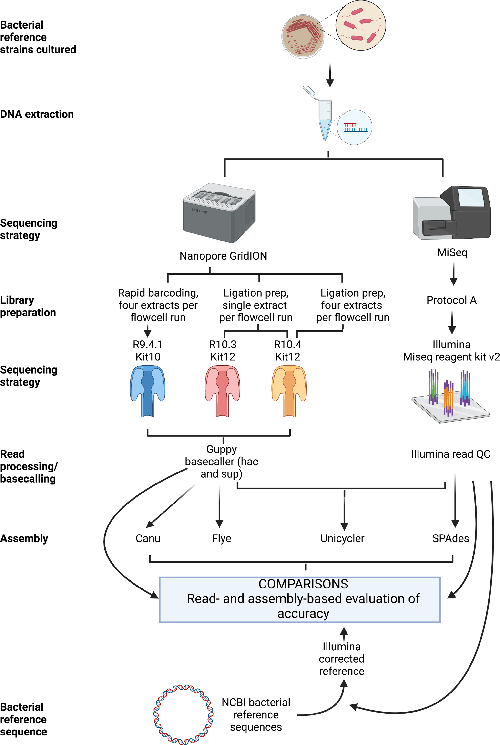
Comparison of R9.4.1/Kit10 and R10/Kit12 Oxford Nanopore flowcells and chemistries in bacterial genome reconstruction | Microbiology Society
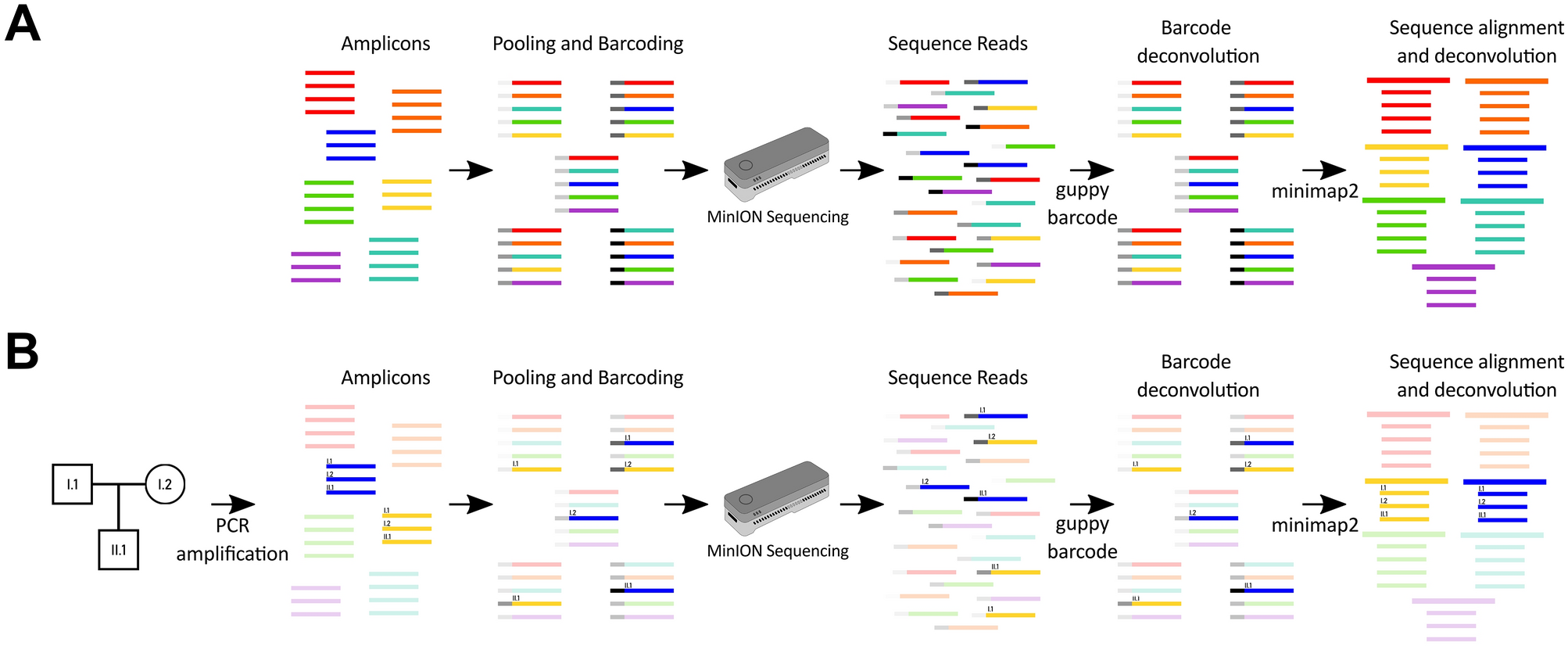
Proof of concept for multiplex amplicon sequencing for mutation identification using the MinION nanopore sequencer | Scientific Reports
GitHub - asadprodhan/GPU-accelerated-guppy-basecalling: GPU-accelerated guppy basecalling and demultiplexing on Linux

Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing | Nature Communications
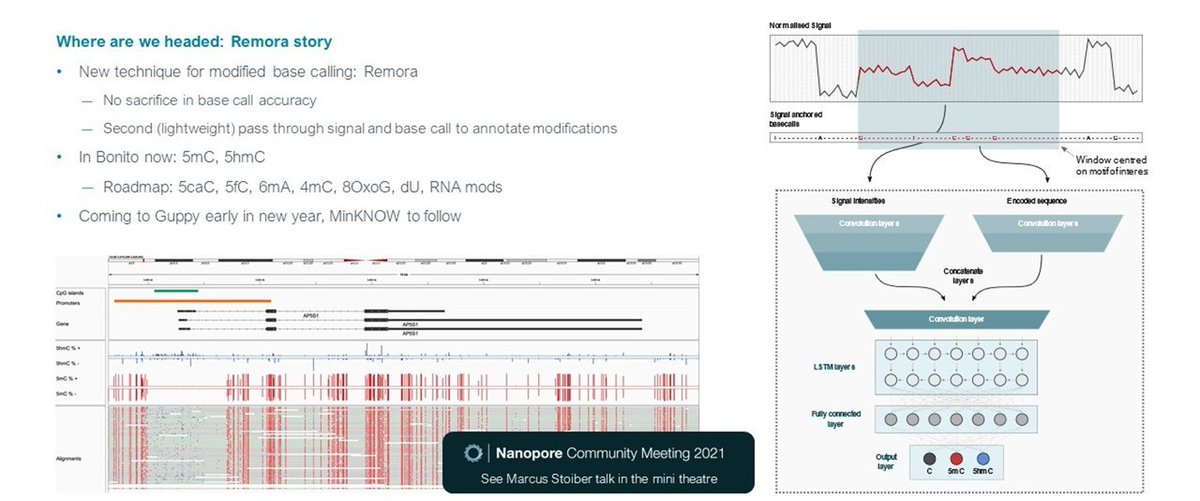
Oxford Nanopore on Twitter: "SR: Remora is 2nd, lightweight pass through the signal, that can run with basecalling, sacrificing no accuracy when delivering methylation analysis in the same experiment. Now in Bonito